
Probabilistic Machine Learning
ProbCMap Software
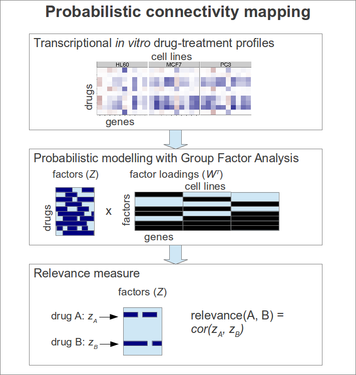
ProbCMap: Probabilistic drug connectivity mapping
Probabilistic drug connectivity mapping can be used to infer drug similarities based on drug treatment gene expression measurements from multiple cell lines, such as the Connectivity Map data. The Group Factor Analysis model is first applied to the expresssion data, separating cell line-specific responses from those that are shared by two or more cell lines. Drug similarities are then computed in the captured model space. In addition to single drug retrieval, also combinatorial retrieval is possible, searching for pairs of drugs that explain the query drug well.
The method has been described in published in Probabilistic drug connectivity mapping. The method has been developed by the Statistical Machine Learning and Bioinformatics group at the Helsinki Institute for Information Technology HIIT, Department of Information and Computer Science, Aalto University.
Citation
When you use this method, please cite the following paper:
Juuso A Parkkinen and Samuel Kaski
Probabilistic drug connectivity mapping
BMC Biofinformatics 2014, 15:113.
Contents
Code and data: ProbCMap_1.0.zip
Contact
Licence
Copyright (C) 2013-2014 Juuso Parkkinen. All rights reserved.
This program is open source software; you can redistribute it and/or modify it under the terms of the FreeBSD License (keep this notice): http://en.wikipedia.org/wiki/BSD_licenses
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE.